Official Journals By StatPerson Publication
|
Table of Content-Volume 5 Issue 3 March 2018
Interspecies communication in oral biofilm
Esha Tyagi*, Pragya Jha, Vikas Kacran, Tanushree Bera, Rinky Tripathi
Department of Periodontics, Army College of Dental Sciences, Rajiv Swagruha ABHIMAAN Project, Secunderabad, Telangana 500083 Email: eshashimmer@gmail.com
Abstract The diversity of signalling opportunities within microbial communities, and the significant role of these molecules in coordinating gene expression and promoting biofilm formation, has provided the impetus to investigate the potential of inhibitory analogues to disrupt these networks, thereby providing mechanisms to control or influence the development of dental plaque. Within the oral biofilms, resident bacterial cells interact with one another and exchange messages in the form of signalling molecules and metabolites. In this review article, our aim is to elaborate on this mutualistic partnership the role of this quorum sensing and their involvement in pathogenesis to decipher information that can be useful to target pathways to control diseases. Key Words: oral biofilm.
The human fetus inside the uterus is sterile but as soon as it passes through the birth canal, it acquires vaginal and fecal microorganisms. Within 2 weeks, a nearly mature microbiota is established in the gut of the newborn baby. Within 2 weeks, a nearly mature microbiota is established in the gut of the newborn baby. After weaning (>2 years), the entire human microbiota is formed and comprises a very complex collection of hundreds of different types of bacteria, totalling approximately 1014 microbial cells. It has been estimated that for a normal, healthy human being, the bacterial population comprises 2 kg of the total body weight. The colonization of the oral cavity also starts close to the time of birth. Communication is a key element in successful organizations. The mouth is similar to other habitats within the body in having a characteristic microbial community that provides benefits for the host. The mouth is warm and moist, and is able to support the growth of a distinctive collection of microorganisms (viruses, mycoplasma, bacteria, Archaea, fungi and protozoa). The bacteria on human teeth and oral mucosa have developed the means by which to communicate and thereby form successful organizations. These bacteria have coevolved with their host to establish a highly sophisticated relationship in which both pathogenic and mutualistic bacteria coexist in homeostasis. The foundations of dental plaque are laid by the primary colonizers, predominantly streptococci, actinomyces and a few other genera.
Table 1: Properties of biofilms and microbial communities (adapted form Ref.90)
Without retention on the tooth surface, the bacteria are swallowed with the saliva. Through retention, these bacteria can form organized, intimate, multispecies communities referred to as dental plaque. Bacteria have often been studied as populations of cells that act independently, but it now seems that there is much interaction and communication between cells. Bacteria can produce an extensive repertoire of secondary metabolites, and can respond to a wide variety of chemicals in their environment. In recent years, particular groups of secondary metabolites have been characterized for their role in the regulation of gene expression in a cell-density-dependent manner, and this behaviour has been collectively referred to as quorum sensing, or cell–cell communication.
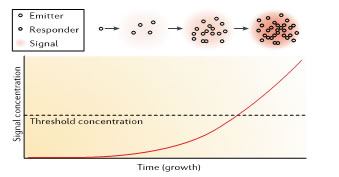
Figure 1: scheme for quorum sensing in its simplest from, cell-cells signalling molecules by emitter cell and their accumulation in the signalling molecules bind to receptors on or in the bacterial cell leading to change in gene expression in the responding cell. For intraspecies quoram sensing, the emitter and responder are usually the same cells, as illustrated here. Often, but not always. The genes that are involved in synthesis and response activate their own expression explaining the term authoinducer. A signalling molecule is considred to act at low concentration and not to he involved in primary metabolism.
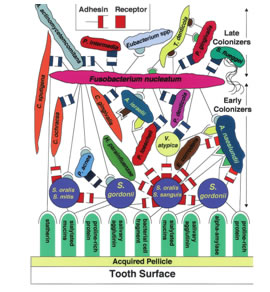
Dental plaque – a classical multi- species biofilm: Dental plaque has been defined as the microbial community that develops on the tooth surface, embedded in a matrix of polymers of bacterial and salivary origin (90). Dental plaque forms via an or- dered sequence of events, resulting in a structurally and functionally organized species-rich microbial biofilm (66, 67, 83, 130). The distinct stages in plaque biofilm formation are described below. Reversible adhesion: Reversible adhesion involves weak, long-range, physico-chemical interactions between the charge on the microbial cell surface and that produced by the conditioning film (8, 19). Microorganisms are usually transported passively to the surface by the flow of saliva or gingival crevicular fluid; a few species (e.g. Wolinella, Selenomonas and Campylobacter spp.) found subgingivally have flagella and are motile. Irreversible adhesion: Irreversible adhesion involves interactions between specific molecules on the microbial cell surface (ad- hesins) and complementary molecules (receptors) present in the acquired pellicle. These adhesin– receptor interactions are strong and operate over a relatively short distance (159), and are targets for possible novel interventions to block colonization. Co-adhesion: During co-adhesion, secondary and late colonizers ad- here via cell-surface adhesins to receptors on already attached bacteria65, leading to an increase in micro- bial diversity within the developing biofilm (microbial succession) (Fig. 3) (67). Many of the secondary colonizers have fastidious growth requirements. Multiplication of the attached cells Multiplication of the attached cells leads to an in- crease in biomass and synthesis of exopolymers to form a biofilm matrix (5, 15). A matrix is a common feature of all biofilms, and is more than a chemical scaffold to maintain the shape of the biofilm. It makes a significant contribution to the structural Plaque as a biofilm and community – consequences for the microorganisms Dental plaque was the first biofilm to be studied in terms of both its microbial composition and its sen- sitivity to antimicrobial agents. It is only in recent years, with the advent and application of new molecular and imaging technologies, that a more complete understanding of the biology of dental plaque as a biofilm and microbial community has been possible. Some of the implications of this surface-associated, community-driven lifestyle, and the opportunities for biofilm control, are described below. SPATIOTEMPORAL MODEL OF ORAL BACTERIAL COLONIZATION Development of the oral microbial community involves com- petition as well as cooperation among the 500 species that compose this community. A few of those oral species are shown in Fig. 1 in a diagram illustrating competition and co- operation among early and late colonizers of the tooth surface. The acquired pellicle, which is composed of a variety of host- derived molecules, coats the enamel surface within minutes after professional cleaning and is a source of receptors recognized by the primary colonizers of dental plaque. These receptors include mucins, agglutinins, proline-rich proteins, phos- phate-rich proteins such as statherin, and enzymes such as alpha-amylase. Each is a known receptor for particular oral species.
Figure 2:
Streptococci constitute 60 to 90% of the bacteria that colo- nize the teeth in the first 4 h after professional cleaning (115). Other early colonizers include Actinomycesspp., Capnocyto- phagaspp., Eikenellaspp., Haemophilusspp., Prevotellaspp., Propionibacterium spp., and Veillonellaspp. The complementary symbols depict physical interactions known to occur between a pair of species. The different shapes and colors of the complementary symbols in represent potentially distinct coaggregations. Rectangle-shaped symbols of any color represent lactose-inhibitablecoaggregations, which are prevalent among oral bacteria Those of the same color represent functionally similar but not identical co- aggregations. The GalNAc133 Gal receptor site in 1 Gn is recognized by functionally similar adhesins on Streptococcus gordonii, Haemophilusparainfluen- zae, Prevotellaloescheii, Veillonellaatypica, Eikenellacorrodens, and Actinomycesnaeslundii. These adhesins are of various molecular sizes, and the species bearing the adhesins compete with each other for binding to the receptor polysaccharide Thus, it is postulated that coaggregation and coad- herence are integral to communication between species and help to establish patterns of spatiotemporal development. Metabolic Communication: The examples of metabolic communication discussed here are limited to interactions in which at least one organism benefits. This arena of metabolic communications among oral bacteria has been reviewed extensively .Beneficial interactions may occur through the excretion of a metabolite by one organism that can be used as a nutrient by a different organism or through the breakdown of a substrate by the extracellular enzymatic activity of one organism that creates biologically available substrates for different organisms. An example of the latter enzymatic activity is sequential hydrolysis of a complex glycoprotein by several bacteria acting in sequence on the product of a previous bacterium’s action, as has been shown for oral streptococci Within the oral cavity, bacteria form multispecies communities that are distinguishable primarily by their location. The subgingival community has the highest species richness and the greatest capacity for pathogenic outcome, such as periodontal tissue destruction. In an examination of cocultures of putative periodontal pathogens, such as P. gingivalisand T. denticola, cocultures produced more biomass than was observed in the respective monocultures; most of the coculture biomass was in the form of cell aggregates, and the coculture was transferable over at least five successive inoculations. MECHANISMS OF COMMUNICATION CONCLUSION AND FUTURE DIRECTIONS Developing oral prophylactic strategies through interference with two-component systems or quorum-sensing of biofilm micro-organisms represents an interesting future challenge. Unlike strategies that target microbial viability, such approach- es may interfere with microbial adaptive pathways without killing the micro-organisms. Therefore, resistance development would probably represent a minor problem. While the stages of biofilm formation seem to follow basi- cally the same model in various micro-organisms, the biofilm architecture and molecular mechanisms involved in biofilm formation appear to differ. The mechanisms involved in biofilm formation by P. aeruginosa are some of the best-charac- terized and have served as a model for new hypotheses on mechanisms used by other micro-organisms. Information on the genetic regulation of oral biofilm formation, however, is still lacking. A better understanding of these processes is nec- essary to the development of novel strategies for oral disease prevention and control based on interference of two-compo- nent signal transduction systems or quorum-sensing. Since the systems contain both conserved and variable components, both broad- and narrow-spectrum responses may be available. This could allow for tailoring of prophylactic measures based on individual oral health status and risk assessment.
REFERENCES
|
 Home
Home