Official Journals By StatPerson Publication
|
Table of Content Volume 10 Issue 1 - April 2019
Annet Olinda Dsouza1, Rajeswarie S2*
1Assistant professor, Department of Microbiology, Kanachur Institute of Medical Sciences and Research Centre, Mangalore, Karnataka, INDIA. 2Assistant Professor, Department of Microbiology, Al-Azhar Medical College and Super Specialty Hospital, Thodupuzha, Kerala, INDIA. Email: annetdsouza13@gmail.com
Abstract Background: Lower respiratory tract infection (LRTI) is one of the commonest encountered disease in the hospital with wide range of disease manifestations like pneumonia, bronchiectasis, acute bronchitis, and acute exacerbation of lung disease. The timely diagnosis and appropriate antibiotic therapy can reduce the morbidity and mortality in a patient. Objectives: This study aims to isolate the pathogens causing LRTI and to study its antibiogram pattern with special reference to ESBL, MBL and Amp C detection which helps in prompt start of antibiotics. Materials and methodology: A total of 55 sputum samples were collected from both inpatient and outpatient departments from August 2018 to September 2018.Out of 55 samples, only 16 samples were poceeded for sputum culture according to Barlett grading’s of sputum sample. Sputum samples were inoculated in blood agar, chocolate agar and Mac conkey agar and further biochemical tests were performed according to the standard protocol.Antibiotic Susceptibility tests was performed along with detection of ESBL, MBL, AmpC and Mehicillin resistance as per standard protocols. Result: Out of 16 samples, Pseudomonas aeruginosa and Klebsiella pneumoniae was isolated in 10 sputum samples accounting for 5 each, 2 were Acinetobacter species followed by 4 isolates each of Citrobacter spp, Escherichia.coli, Streptococcus pneumoniae and Staphylococcus aureus. 40% of the Klebsiella pneumoniae isolates produced ESBL and 20% produced AmpC. The one isolate of Citrobacter spp produced both ESBL and AmpC. 60% of the Pseudomonas aeruginosa and 50% of the Acinetobacter spp produced MBL. Key Word:Antibiotic sensitivity, Bartlett grading, Lower respiratory tract infection, Multi drug resistant bacteria. INTRODUCTION The most common infectious disease encountered in the hospital is the Lower Respiratory Tract Infections (LRTI) and these infections have emerged as a leading cause of morbidity and mortality1.LRTI is a broad description of a variety of diseases of the lower respiratory tract consisting of the acute bronchitis, pneumonia, bronchiectasis and acute exacerbation of chronic lung disease2. Determining the exact pathogen has been confounded because of the presence of the commensal flora as well as of the pathogenic organisms in the oropharynx. With the increasing antibiotic resistance in the bacterial pathogens causing the respiratory tract infections it has been a great challenge to start the empiric treatment. Infections with the multi drug resistant organisms leads to poor outcome, prolongs the hospital stay and increases the hospital costs, and increases the risk of morbidity and mortality30.Monitoring of the resistance pattern of the bacteria not only assists the clinicians but also helps in surveillance of LRTI. The inappropriate treatment and abuse of antimicrobial drugs has often given rise to resistance to the most commonly used antibiotics4.Multidrug resistant organisms may be associated with the illness or may lead to asymptomatic carriage. Information on LRTI by multi drug resistant organisms is limited. The purpose of this prospective study was to investigate the bacterial agents responsible for LRTI in a tertiary care hospital setting and study their antibiogram so as to guide in initiating empirical therapy to reduce the mortality and morbidity associated with it.
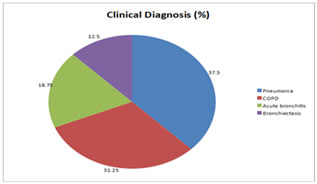
MATERIALS AND METHODS This was a descriptive study in which the sputum samples were collected from both out and in patients of a tertiary care hospital and was conducted in the Department of Microbiology, Kanachur Institute of Medical Sciences from August 2018 to September 2018. All adult patients above 18 years of age with clinical signs and features suggestive of LRTI on clinical examination and radiological signs suggestive of LRTI were included in the study. Those patients who were on antibiotics for more than 48 hours were excluded from the study. The sputum samples were assessed for the quality using the Bartlett scoring system. The sputum sample was subjected to Gram stain to look for the presence of leucocytes, squamous epithelial cells and bacteria. As per the scoring system the sputum samples were scored as 0, +1, or +2 as per the number of leucocytes seen per low power field, and 0, -1, and -2 as per the number of squamous epithelial cells seen per low power field. The samples with a score of 0 or less were not considered for the study as it was contaminated with the oro pharyngeal flora. Only the sputum samples with more than 25 leucocytes and less than 10 squamous epithelial cells per low power field were considered as an ideal quality sample and were further processed for culture and antimicrobial sensitivity testing 5.The sputum samples were inoculated on 5% sheep blood agar, chocolate agar and MacConkey agar and incubated at 37OC for 18 to 24 hours. From the isolated colonies, Gram stain was performed and the isolate was identified by biochemical reactions as per the standard protocol for identification9. Antibiotic susceptibility testing was done for the isolates using Kirby-Bauer’s disc diffusion technique on Mueller-Hinton agar (BD) as per the Clinical Laboratory Standard’s Institute guidelines10. The isolates obtained were then checked for extended spectrum beta lactamase (ESBL) by double disc approximation, Amp C β lactamase by disc antagonism and Metallo β-lactamase detection by EDTA disc synergy test11. All statistical analysis was performed by using a statistical Software program (SPSS version 22). Frequency tables and graphs were produced for the data obtained. RESULTS Majority of the patients 6 (37.5%) were clinically diagnosed as case of pneumonia, 5(31.25%) had acute bronchitis and about 2 (21.5%) had bronchiectasis. (Figure 1) Figure 1: Clinical diagnosis of study population with LRTI
There were total of 16 bacterial isolates obtained. Of them 14 were Gram negative isolates, 5(35.7%) Pseudomonas aeruginosa and 5(35.7%) Klebsiella pneumoniae were the predominant isolates. Among the 2 Gram positive cocci isolates, one was Streptococcus pneumoniae and the other was Methicillin Sensitive Staphylococcus aureus as seen in table 1.
Table 1: Organisms isolated in LRTI patients
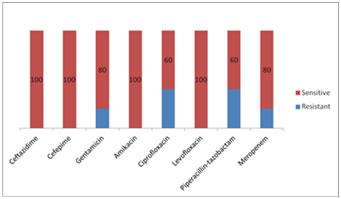
Figure 3: Antibiotic sensitivity pattern in Pseudomonas aeruginosa
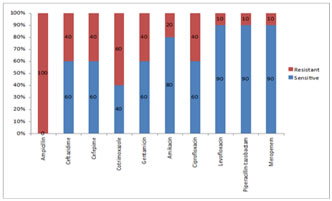
Of the Klebsiella pneumoniae isolates 60% were resistant to Cotrimoxazole and there was resistance seen to the Beta lactam- Beta lactamase inhibitor combinations as seen in figure 2. The Pseudomonas aeruginosa isolates were resistant to Ciprofloxacin in 40% and 40% were resistant to Piperacillin-tazobactam.
DISCUSSION During the study period from August 2018 to September 2018, a total of 55 cases of Lower Respiratory Tract Infections (LRTI) were recruited in the study. All the 55 sputum samples were screened by Gram stain and scored as per the Bartlett grading system. 38(69.09%) of the samples fulfilled the criteria of a good quality sputum sample. Only 16 samples showed growth and they were confirmed by microbiological culture and sensitivity. Of the 16 bacterial isolates, 14(87.5%) were Gram negative bacilli and 02(12.5%) were Gram positive cocci. The mean age group of the population was 52.68years. This is in par with other studies from India6. The study comprised of 9(56.25%) males and 7(43.75%) females. Majority of the population were males, in whom smoking was the common risk factor. The risk factors involved in our study with LRTI were smoking in 6(37%), diabetes mellitus in 4(25 %), seasonal variation in 3(19%) and unknown in 3(19%). In this study, it was predominantly monomicrobial with only one organism grown in culture. The monomicrobial growth was obtained in 15(93.75%) samples and 01(6.25%) had polymicrobial growth. The polymicrobial growth constituted of Citrobacter spp. and Candida albicans. Polymicrobial growth of Citrobacter spp. and Candida albicans was seen in an ICU patient with pneumonia. Of the 16 cases in our study, 6(37.5%) were patients with pneumonia, 5(31.25%) were acute exacerbation of COPD, 3(18.25%) were patients with acute bronchiectasis and 2(12.5%) were patients with acute bronchitis. Majority of the patients presented with clinical findings and chest X-Ray findings of pneumonia. All these cases were clinically suspected and diagnosis was confirmed by microbiological examination of sputum samples. In the study, a total of 14(87.5%) cases had Gram negative bacilli and 2(12.5%) had Gram positive cocci grown in culture. Of the 14 cases, 5(35.7%) were Pseudomonas aeruginosa and 5(35.7%) were Klebsiella pneumoniae and these topped the list of microorganisms isolated from LRTI. The other Gram negative bacteria isolated were Acinetobacter spp, Pseudomonas aerginosa in 2(14.26%), E.coli in 1(7.1%) and Citrobacter spp in 2(14.28%). The sputum samples with Gram positive cocci 2(12.5%) had growth of MSSA in 1(50%) and Streptococcus pneumoniae in 1(50%). Of the 6 cases of pneumonia, Klebsiella pneumoniae was isolated from 3(50%) of the cases, Acinetobacter species in 2(33.3%), and Pseudomonas aeruginosa in 1(16.6%). Among the 5 cases of acute exacerbation of COPD, 5 isolates were obtained. There was 1(20%) isolate each of Pseudomonas aeruginosa, Klebsiella pneumoniae, E.coli, Streptococcus pneumoniae and MSSA. Both the Gram positive cocci were from patients with acute exacerbation of COPD. In the 3 patients of acute bronchiectasis, the isolates obtained were Pseudomonas aeruginosa 1(33.3%), Klebsiella pneumoniae 1(33.3%) and Citrobacter species 1(33.3%). Acute bronchitis was seen in 2 cases. The isolates obtained were of Pseudomonas aeruginosa in 2(100%). In a study done by Khan S et al 7the common organisms isolated were, Pseudomonas aeruginosa in 71(35.34%) and Klebsiella pneumoniae in 68(33.53%) of patients. The same holds good in our study also, where Pseudomonas aeruginosa and Klebsiella pneumoniae were the common organisms of isolated. It is of concern that Pseudomonas aeruginosa is an example of opportunistic bacterial pathogen and is well reported as nosocomial infection. Klebsiella pneumoniae accounts for LRTI in India. Gram negative bacilli- The isolates obtained were E.coli, Klebsiella pneumoniae, Pseudomonas, Acinetobacter and Citrobacter. The 1 E.coli isolate was a sensitive strain with sensitivity to all the classes of antibiotics. Of the 5 isolates of Klebsiella pneumoniae there was resistance seen in 3(60%) of isolates to Cotrimoxazole, 1(20%) to Amikacin, 2(40%) to Ciprofloxacin and 2nd and 3rd generation cephalosporins. The 5 isolates of Pseudomonas aeruginosa were sensitive to 2(40%) Ciprofloxacin and 1(10%) to Levofloxacin and Piperacillin-Tazobactum, 1(10%) to Meropenem. The isolate Citrobacter spp was totally resistant to all group of antibiotics. Gram positive cocci-Of the 2 Gram positive cocci isolated, one isolate was MSSA and the other was Streptococcus pneumoniae. Both of them were isolated from COPD patients. The strains were sensitive to the antibiotics tested. Drug resistance mechanisms- Among the 07 Enterobacteriaceae isolates, some produced ESBL and AmpC enzymes. Of the 5 Klebsiella pneumoniae isolated, 2(40%) were ESBL enzyme producers, among that 1(20%) produced AmpC. The Citrobacter 1(100%) produced ESBL and AmpC. There were a total of 7 non fermenting Gram negative bacilli, of which 5 were Pseudomonas aeruginosa and 2 were Acinetobacter species. Of the 5 Pseudomonas aeruginosa, 3(60%) produced MBL and of the 2 Acinetobacter spp 1(50%) produced MBL. Similar findings of drug resistance strains are seen in various studies done in India.12
CONCLUSION The high resistance rate to various antimicrobials observed in the present study could be attributed to the fact that LRTI is commonly treated by general practitioners without culture and sensitivity report. There is use and misuse of the broad spectrum antibiotics that has given rise to this problem. The lack of a definite antibiotic policy is a matter of great concern in management of LRTI. To conclude, this study suggests that the most common organism isolated in lower respiratory tract infections is Klebsiella pneumoniae, Pseudomonas aeruginosa and Streptococcus pneumoniae among Gram negative bacilli and Gram positive cocci respectively. By knowing the antibiotic sensitivity pattern of these organisms antibiotic sensitivity policy for the institution can be drafted.
REFERENCES
|
|
 Home
Home