Official Journals By StatPerson Publication
|
Table of Content Volume 11 Issue 2 - August 2019
Evaluation of inducible clindamycin resistance among staphylococcus aureus isolates in a tertiary care hospital
Vijaya Durga Suryadevara1*, Anuradha Basavaraju2
1Associate Professor, Professor and HOD, Department of Microbiology, Mamata Medical College, Khammam, INDIA. Email: durgasv11@yahoo.com
Abstract Background: Macrolide resistance in staphylococci may be due to efflux encoded by msrA gene or ribosomal target modification [macrolide lincosamide streptogramiun B (MLSB) resistance], encoded by erm A or erm C genes. Following exposure to a macrolide, MLSB resistance is either constitutive or inducible. Aim: This study was aimed to detect the presence of inducible Clindamycin resistance among clinical isolates of Staphylococcus aureus using ‘D – test’. Materials and Methods: A total of 265 S. aureus isolates were evaluated and methicillin – resistance was determined using Cefoxitin (30µg) disc. Inducible Clindamycin resistance was detected by ‘D–zone’ test as per CLSI guidelines on erythromycin resistance isolates. Results: Of the 265 Staphylococcus aureus strains 87 (32.83%) were identified as MRSA and 178 (67.17%) as MSSA. Sixty one (23.01%) isolates showed inducible Clindamycin resistance (iMLSB), 21 (7.92%) showed constitutive resistance (cMLSB) and 24 (9.05%) were erythromycin resistant and clindamycin sensitive strains (MSB) phenotype). Remaining 159 (68.57%) were sensitive to both clindamycin and erythromycin. Inducible and constitutive phenotypes were found higher in MRSA compared to MSSA. Conclusion: Prevalence of Clindamycin resistance is higher in MRSA isolates as compared to MSSA isolates. Routine D– test should be included in the routine antibiotic susceptibility testing as it will guide the clinician about iMLSB phenotype of Staphylococcus aureus, so that Clindamycin may be used judiciously. Key Word: macrolode lincosamide streptogramin B phenotype, Clindamycin, MS phenotype, Staphylococcus aureus.
INTRODUCTION Staphylococcus aureus produces various infections, from minor skin infection to life threatening infections1. MRSA incidence in India ranges from 30 – 70%2. The increasing frequency of methycillin resistant Staphylococcus aureus (MRSA) infections and changing patterns in antimicrobial resistance have led to renewed interest in the usage of macrolide- lincosamide-streptogramin B (MLSB) antibiotics to treat such infections with Clindamycin being the preferable agent due to its excellent pharmacokinetic properties3. However their wide spread use has led to increase in number of staphylococcal strains developing resistance to MLSB antibiotics4. The most common mechanism for such resistance is target site modification mediated by erm genes which can be expressed either inducibly (iMLSB phenotype) or constitutively (cMLSB phenotype) and also develop isolated macrolide resistance based on presence of an efflux pump, encoded by msrA gene which leads to resistance to macrolides and type B streptogramins but not to lincosamides5, these isolates are MS phenotype which also show invitro resistance to Erythomycin (ER) and susceptibility to Clindamycin (CD) same as in inducible resistance phenotype, but CD therapy can be safely given in infections with this phenotype and there is no risk of clinical failure as in iMLSB phenotype. Therefore it is important to differentiate these three mechanisms of resistance. Unfortunately the iMLSB phenotype cannot be recognized using standard susceptibility methods including standard broth based or agar dilution tests. Low levels of ER is an inducer of iMLSB phenotype, which forms the basis of D – test4. Phenotypic detection of inducible resistance can be done by double disk diffusion test (D – test) as describe by Fiebelkorn and co-workers5, in which distorted ‘D – shaped’ zone of inhibition is observed around CD if an ER disc is placed nearby (15mm)6. The present study was undertaken to determine the prevalence of resistance to erythromycin and Clindamycin in S. aureus isolated from various clinical samples in a tertiary care hospital to assist clinicians in the treatment of these infections by these group of antibiotics. The study was aimed to determine the prevalence of inducible Clindamycin resistance (iMLSB), constitutive (cMLSB) and MSB phenotype in isolates of Staphylococcus aureus in our geographical area using D – test and to ascertain the relationship between MRSA and iMLSB, cMLSB and MSB resistance.
MATERIALS and METHODS The present prospective cohort study was conducted in a tertiary care hospital from February 2013 to December 2016 after obtaining institutional ethical committee clearance and informed consent from participants. Statistical analysis was done by chi square test using SPSS soft ware version 2.1. Our study included 265 non-duplicate Staphylococcus aureus isolates from various samples of pus, sputum, blood, urine and body fluids from patients of both admitted cases (IPD) and outpatient departments (OPD). Staphylococcus aureus isolates were identified by using standard microbiological procedures (gram staining, culture, catalase test, slide and tube coagulase test, mannitol fermentation and production of DNAase enzyme)7. Antibiotic sensitivity testing of all the strains were performed by modified kirby Bauer disc diffusion method as per CLSI guidelines for the following antibiotics penicillin (10units), gentamycin (10µg), Erythromycin (15 µg), ciprofloxacin (5 µg), Clindamycin (2µg), tetracycline (30µg), amoxycillin/clavulanic acid (20/10 µg), trimethoprim/sulfamethoxyzole (1.25/23.75µg), Cefoxitin (30µg) linezolid (30µg) and Vancomycin (30µg). Methicillin resistance in Staphylococcus aureus was detected by using 30µg Cefoxitin disc8. Staphylococcus aureus ATCC 25923 was used as the control strain for the disc diffusion method. S. aureus isolates which were resistant to erythromycin(ER) and Clindamycin (CD) sensitive were further tested by D-zone test for finding inducible Clindamycin resistance as per CLSI guidelines, by inoculating bacterial suspension adjusted to 0.5 Mc farland’s standard on muller– hinton agar plate and placing erythromycin(15µg) and Clindamycin(2µg) disks side by side with edge to edge distance of 15mm8. Plates were analysed after overnight incubation at 370 C. Appearance of CD disc zone close to ER disc was noted and three different phenotypes were observed after testing and interpreted as follows -
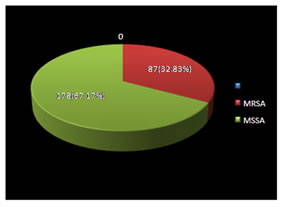
RESULTS Among the 265 Staphylococcus aureus isolates 87 (32.83%) were methicillin resistant Staphylococcus aureus (MRSA) and 178 (67.17%) were methicillin sensitive Staphylococcus aureus (Figure1). Figure 1: Prevalence of MRSA and MSSA strains (N = 265)
Table 1: Prevalence of iMLSB, cMLSB and MSB phenotypes in MRSA and MSSA isolates
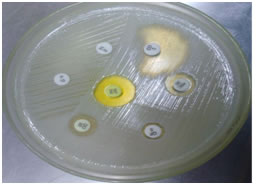
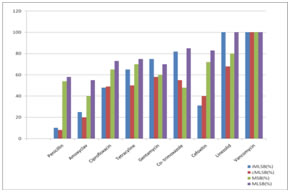
Table 1 shows, out of 265 Staphylococcus aureus isolates 159 (68.57%) were sensitive to both ER and CD (MLSB), 21(7.92%) were resistant to both ER and CD (cMLSB phenotype), 61(23.01%) were D-test positive (iMLSB phenotype) (Figure 2) and 24 (9.05%) were D-test negative (MSB phenotype). Resistance percentage was high in both iMLSB phenotype and cMLSB phenotype among MRSA isolates (45.97% and 13.79% respectively) as compared to MSSA isolates (11.79% and 5.05% respectively). High numbers of MSSA isolates (73.03%) were sensitive to both ER and CD than MRSA isolates (33.33%). This finding was found to be insignificant (P>0.05). The antibiotic sensitivity pattern of all the four phenotypes reveal that they were 100% sensitive to Vancomycin and least sensitive to penicillin (Figure 3).
Figure 2: Inducible Clindamycin resistance Staphylococcus aureus (iMLSB phenotype) Figure 3: Antibiotic susceptibility profile of inducible MLSB, constitutive MLSB, MSB and MLSB Staphylococci aureus strains DISCUSSION For optimal antimicrobial therapy of infected patients, determination of antimicrobial susceptibility of a clinical isolate is often crucial. This is particularly important considering the increase of resistance and the emergence of multidrug resistant organisms. There are many options available for treatment of MRSA and MSSA, among which Clindamycin being one of the good alternatives6. However, Clindamycin resistance can develop in staphylococcal isolates, differentiation of ‘erm’ mediated inducible MLSB (iMLSB phenotype) from isolates with msr A mediated(MSB) phenotype is a critical issue for any clinical laboratory because of the therapeutic implications of using clindamycin and erythromycin to treat patients with an inducible Clindamycin resistant Staphylococcus aureus isolates9. Clindamycin is an excellent drug for treating skin and soft tissue infections caused by Staphylococcus aureus and are also an alternative in penicillin–allergic patients and in infections due to MRSA10. Clindamycin has good oral bioavailability making it good option for outpatient therapy and changeover after intravenous antibiotics. However, Clindamycin resistance in inducible phenotype can lead to development of spontaneous constitutively resistant mutants both invitro and invivo during Clindamycin therapy4. In the present study, 265 Staphylococcus aureus isolates of various clinical samples of both outpatient departments (OPD) and admitted cases (IPD) were included. The prevalence of MRSA was found to be 32.83% which correlates with our previous study 32.12%11. Prevalence of MRSA varies in different areas from 0.4 to 48.4%. In our study, of the 87(32.83%) MRSA isolates, 40(45.97%) were of iMLSB phenotype and 12 (13.79%) were cMLSB phenotype where as of the 173 (67.17%) MSSA isolates 21 (11.79%) were iMLSB and 9 (5.05%) were cMLSB phenotype. It was observed that inducible resistance was much higher in both MRSA and MSSA isolates. Of the 106(40%) isolates irrespective of whether MRSA or MSSA they were resistant to Erythromycin by routine disc diffusion testing. Among them 61 (57.5%) showed D – test positive, indicating iMLSB while 24 isolates (22.64%) showed negative D – test and were truly sensitive to Clindamycin indicating MSB phenotype and whereas 21 (19.81%) were resistant to both erythromycin and Clindamycin indicating cMLSB. These observations suggest that, if D – test had not been performed, more than half of the erythromycin resistant isolates would have been misidentified as Clindamycin sensitive resulting in therapentic failure. Conversely, labeling all ER–resistant Staphylococcus aureus as CD – resistant prevents the use of CD in infections caused by truly CD sensitive staphylococcal strains. In present study we also looked forward for treatment options for iMLSB isolates by detecting their antimicrobial sensitivity to other antibiotics it was found that all the isolates of iMLSB phenotype were 100% susceptible to linezolid and vancomycin followed by moderate susceptibility to co–trimoxazole and gentamycin and least susceptibility to amoxyclav and ciprofloxacin. Our findings are in correlation to other studies that also found all iMLSB isolates were uniformly susceptible to linezolid and Vancomycin12-15. significantly higher resistance was exhibited by CMLSB towards amoxyclav and ciprofloxacin and MSB phenotype towards amoxyclav only.
CONCLUSION Clindamycin is one of the most commonly used antibiotics for MRSA as well as MSSA. The increasing Clindamycin resistance in the form of inducible and constitutive MLSB limits the therapeutic option. The true sensitivity of Clindamycin can only be judged after performing D- test on erythromycin resistant isolates. The implementation of D– test in routine antibiotic susceptibility testing enable to delineate inducible and constitutive Clindamycin resistance. Therefore as recommended by CLSI, D – zone test should be routinely performed in all laboratories to guide the clinicians regarding judicious use of Clindamycin in skin and soft tissue infections.
ACKNOWLEDGEMENT We wish to express our profound gratitude to all patients who participated in this research study.
REFERENCES
|
|
 Home
Home