Official Journals By StatPerson Publication
|
Table of Content Volume 12 Issue 1 - October 2019
Clinico - microbiological study of blood stream infections among patients admitted to critical care units
Bewin Oral J1*, Madhuri Kulkarni2, Goldy S J3
1Assistant Professor, Department of Microbiology, Dr Somervell Memorial CSI Medical College, Karakonam, Thiruvananthapuram 2Professor, Department of Microbiology, JSS medical college, Mysore. 3Assistant Professor, Department of Obstetrics and Gynecology, Dr Somervell Memorial CSI Medical College, karakonam, Thiruvananthapuram Email: bewin.grace@gmail.com
Abstract Background: Bloodstream infections (BSIs) mostly lead to life threatening sepsis and are associated with high morbidity and mortality. BSIs require immediate and appropriate antimicrobial treatment, since their prevalence, aetiology, and antimicrobial susceptibilities differ from one country to other. Hence Blood culture is the gold standard test providing information for the evaluation of a variety of diseases like endocarditis, pneumonia, and pyrexia of unknown origin particularly, in patients with suspected sepsis, yet their positivity rate is around 19%. This scenario establishes a necessity for conducting my study which can help in forming a protocol in starting an empirical treatment and to have an antibiotic stewardship programme in cases of sepsis. Methods: This study was conducted in JSS hospital, Mysore, South India among the adult patients (>18 yrs.) with clinical signs of sepsis between December 2013 and November 2014. Results: A Total of 100 clinically suspected cases of BSI samples were processed by Bact-Alert system which yielded a culture positivity of 19%. The culture positive samples were identified and the Antimicrobial susceptibility patterns of each organism were obtained using Vitek 2 automated system, which yielded Gram negative bacilli (63%), Gram positive cocci (27%) and Yeast (10%). Escherichia coli (27%), Klebsiella pneumoniae (11%), Acinetobacter baumannii (11%), Staphylococcus aureus (10%), CONS (Staphylococcus epidermidis and Staphylococcus hominis) (10%) were the most frequently obtained isolates from adult septicemia cases. Many of these isolates were multidrug resistant (MDR). Genito-urinary infections (79%), followed by respiratory infections (53%) were the most common source for secondary bacteremia. Conclusion: Blood stream infection and Antimicrobial resistance, particularly among Gram-negative organisms, continues to increase at a rapid rate, especially in the Intensive care units. Coordinated infection control interventions and Antimicrobial stewardship policies are required to lower the emergence of resistance. Key Words: Blood stream infections (BSI), Multi drug resistant (MDR), Sepsis, Antimicrobial susceptibility.
INTRODUCTION Blood stream infection (BSI) is a severe life-threatening condition, especially in critically ill patients, resulting from invasion of blood stream by microbes and consequent clinical manifestation. When bacteria are introduced directly into the circulatory system, especially in a person who is ill or undergoing aggressive medical treatment, the immune system may not be able to cope with the invasion, and symptoms of blood stream infections may develop.1,2,4 In recent years, incidence of BSI in patients admitted to intensive care units (ICU) has increased due to increased use of invasive devices and immunosuppressive therapy. Presently, nosocomial BSI has been reported to be among the most frequently encountered nosocomial infections in the ICU. According to an estimate, community-acquired BSI accounts for 20% of all ICU admissions and 28% of all BSI diagnosed in the ICU. Besides increasing incidence, BSIs also have shown increase in treatment costs, length of stay and mortality. The case-fatality rate from sepsis causing organ dysfunction ranges from 30% to 50%, and mortality up to 35% has been associated with BSI. 2Despite advances in therapy and supportive care, Blood Stream Infection (BSI) continues to be a major cause of morbidity and mortality in hospitalized patients. Intensive care units are often the epicentre of these infections, mainly because of its extremely vulnerable population and the increased risk of becoming infected through multiple invasive diagnostic and therapeutic procedures. Accompanying the physiologic stress of infections is the increasingly added burden of multidrug resistance that hampers therapy of these infections, with consequent adverse clinical and economic results. The on-going emergence of resistance in the community and hospitals is a major threat for the public health system.1Urinary tract infection, intravascular device related BSI, gastrointestinal related blood stream infections, and respiratory tract infections are the most frequent sources of health care associated BSI. Intravascular device related blood stream infections occur with similar frequencies in patients with health care associated BSI and in those with hospital acquired BSI.2 Hence the enhanced and timely detection of blood stream infections is integral to optimum health care, especially in intensive care set up.3Therefore early diagnosis and appropriate treatment of these infections can make the difference between life and death.4Though blood stream infections are a major concern nonetheless they are very much preventable with appropriate knowledge and intervention, supported by clinical data and thus arises the need for such a study. This study is intended to find out the microbiological and clinical profile of BSI in these patients and detecting the sources if any and extend the results of the research in defining protocols for the prevention and control of these common yet very important infections that are a major threat to present day health care setup. AIMS AND OBJECTIVES OF THE STUDY
Materials and Methods Source of data: The present study was conducted in the department of Microbiology, JSS tertiary care hospital. The study was carried out from December 2013 to November 2014. Selection of subject: Inclusion criteria: Patients above 18 years of age and having at least two of the clinical signs or manifestations of systemic inflammatory response syndrome and or sepsis were included (High or low temperature, Increase heart rate, Increase respiratory rate, leukopenia or leucocytosis). Exclusion criteria: Patients admitted to intensive care units for short term post-operative monitoring were excluded. Blood Culture Preparation of site and collection of blood 23
Interpretation: Those indicated positive (+) by BacT / Alert 3D system were sub cultured on blood agar and Mac Conkey agar. The blood agar and Mac Conkey agar were incubated at 37 degree Celsius. Various organisms were identified on the basis of colony morphology and standard biochemical tests.23 Those indicated as negative by 7 days (as per setting of Bact / Alert 3D system) were reported as “no growth”. Based on the growth of organisms on primary culture plates; Gram stain and basic standard tests(like Gram stain, catalase, oxidase, hanging drop method) were done according to the Standard operating Protocol and processed with their corresponding vitek 2 identification cards(ID) like Gram Positive, Gram Negative and Yeast ID cards for the further identification to species level and Antimicrobial Susceptibility Testing (AST) with N280 and N281 analysed in the system according to CLSI 2013 guidelines. Identification and the antimicrobial susceptibility patterns of the organisms were obtained from vitek 2 automated system.
Results In present study, blood culture samples from a total of 100 patients above 18years of age and having at least two of the clinical signs or manifestations of systemic inflammatory response syndrome and or sepsis (High or low temperature, Increase heart rate, Increase respiratory rate, leucopenia or leukocytosis) were processed by BacT-Alert system and positive samples were identified and antibiotic susceptibility testing done by vitek 2 automated system.
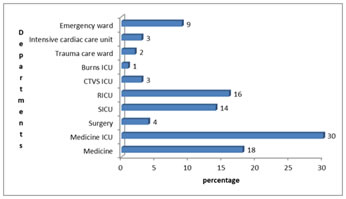
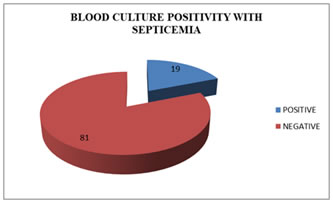
Figure 1: Specialty wise distribution of blood culture samples Figure 2: Blood culture positivity in adults with septicemia Fig 1: Maximum number of samples were from medicine ICU (30%), medicine ward(18%) followed by Respiratory ICU (16%), Surgery ICU (14%) then Emergency ward (9%), Surgery (4%), ICCU (3%) and Trauma care ward (2%). Out of the total 100 blood samples received, 64 (64%) were from males and 36 (36%) were from females. Maximum numbers of samples were from patients in the age group of 46–60 years, accounting for 37%, which was followed by 61-75 years age group- 24%. Fig 2: shows that, out of 100 clinically suspected adult septicemia cases, blood culture was positive in 19 (19%) cases whereas in 81 (81%) cases blood culture was negative. Table 1: Microbial isolates from blood culture (n=100)
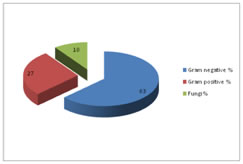
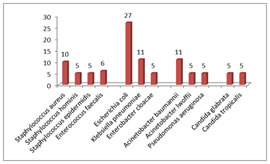
Figure 3: Distribution of culture positive samples based on gram reaction Figure 4: Microbial profile in adult septicemia in blood culture In present study (Fig 3), Gram negative bacilli were found to be the commonest cause of adult septicemia (63%). Gram positive cocci were found in 27% of cases, while Candida glabrata and Candida tropicalis were isolated in 10% of cases. Among Gram negative isolates, Escherichia coli (27%), Klebsiella pneumoniae (11%) were commonest followed by, Acinetobacter baumannii (11%), Acinetobacter lwoffii (5%), Pseudomonas aeruginosa (5%) and Enterobacter cloacae (5%).Among Gram positive isolates, Staphylococcus aureus (10%) was the commonest followed by CoNS (Staphylococcus epidermidis and Staphylococcus hominis) (10%) and Enterococcus faecalis (6%).Among fungi, Candida glabrata (5%) and Candida tropicalis (5%). (Fig 4) Table 2: Distribution of cases based on clinical diagnosis
Genito-urinary infections (79%) followed by respiratory infections (53%), metabolic infections (53%), cardiovascular system infections (53%) and skin and soft tissue infections (11%) were the common clinically suspected primary sources of infection.
Table 3: Antimicrobial sensitivity of Gram positive organisms
P-Penicillin G, OX-Oxacillin, G- gentamycin, Cip – Ciprofloxacin, Levo- Levofloxacin, E- Erythromycin, CD- clindamycin, LZ- linezolid, Dapto- Daptomycin, Teico- teicoplanin, VA- vancomycin, TE- tetracycline, Tgc- Tigecycline, Rif- Rifampicin, Cot- cotrimoxazole, HLG- high level gentamycin. Table 3 shows the antimicrobial sensitivity of Gram positive cocci isolates from blood culture of adult septicemia cases. It shows that Gram positive cocci were 100% sensitive to vancomycin, teicoplanin and linezolid. Two isolates of Staphylococcus aureus showed sensitivity to gentamycin, ciprofloxacin, levofloxacin, clindamycin, linezolid, daptomycin, teicoplanin, vancomycin, tetracycline, tigecycline and rifampicin. Two isolates of Coagulase negative staphylococcus were sensitive to linezolid, teicoplanin, vancomycin, tigecycline and resistant to penicillin, oxacillin, clindamycin and tetracycline. One isolate of Enterococcus faecalis was sensitive to linezolid, teicoplanin, vancomycin, tigecycline and high level gentamycin. Table 4: Antimicrobial sensitivity of Enterobacteriacae isolates (n=8)
A- Ampicillin, AC- Amoxicillin/clavulanic acid, PIT- piperacillin/tazobactam, Cu- cefuroxime, CuA- Cefuroxime axetil, Ctr- Ceftriaxone, Cfs- Cefoperazone/ sulbactam, Cfp- Cefepime, Ert- Ertapenem, Imp- Imipenem, Mrp- Meropenem, Ak- Amikacin, G- Gentamycin, Cip- Ciprofloxacin, Tgc- Tigecycline, Cl- colistin, Cot- cotrimoxazole. All five Escherichia coli isolates were sensitive to ertapenem, meropenem and colistin. Four E.coli isolates were sensitive to Cefoperazone/ sulbactam, amikacin and tigecycline. Three Ecoli isolates were sensitive to piperacillin/tazobactam, imipenem and gentamycin. The two isolates of Klebsiella pneumoniae were sensitive to ertapenem. One of the isolates were sensitive to piperacillin/tazobactam, Cefoperazone/ sulbactam, cefepime, imipenem, meropenem, amikacin, gentamycin, tigecycline and colistin and the other was resistant to these antibiotics. Both isolates were resistant to ampicillin, amoxicillin/ clavulanic acid, cefuroxime, cefuroxime axetil, ceftriaxone and cotrimoxazole. One isolate of Enterobacter cloacae isolated was sensitive to ceftriaxone, Cefoperazone/ sulbactam, cefepime, ertapenem, meropenem, amikacin, gentamycin, ciprofloxacin, tigecycline, colistin and cotrimoxazole and resistant to ampicillin, amoxicillin/clavulanic acid, cefuroxime and cefuroxime axetil.
Table 5: Antimicrobial sensitivity of Pseudomonas aeruginosa and Acinetobacter spp
PIT- piperacillin/tazobactam, CAZ- Ceftazidime, CFS- Cefoperazone/sulbactam, CFP- cefepime, AZT- aztreonam, DORI- doripenem, Imp- imipenem, Mrp- meropenem, G- gentamycin, Cip- ciprofloxacin, Levo- levofloxacin, Mino- minocycline, Tgc- tigecycline, CL- colistin, Cot- cotrimoxazole. Table 5 shows the antimicrobial sensitivity of Pseudomonas aeruginosa. The isolate showed sensitivity to cefepime, doripenem, meropenem, gentamycin, ciprofloxacin, levofloxacin and colistin. The organism was resistant to piperacillin/ tazobactam, ceftazidime, Cefoperazone/ sulbactam, aztreonam, imipenem, minocycline, tigecycline and cotrimoxazole. All three isolates of Acinetobacter spp were sensitive to colistin. Among three isolates two were sensitive to minocycline and tigecycline. One of the three isolates was sensitive to all drugs. Table 6: Antifungal susceptibility pattern of Candida spp
Table 6 shows two isolates of candida spp. with sensitivity to fluconazole, voriconazole, micafungin and flucytosine. Candida glabrata was resistant to caspofungin. Candida tropicalis was resistant to Amphotericin B.
Table 7: Risk factors for sepsis
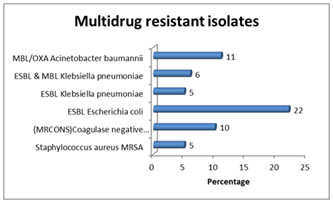
Table 7 shows the risk factors for sepsis: Diabetes mellitus (37% ), intravascular catheter (32% ) , ventilator associated pneumonia (16%), carcinoma (11%) , dialysis (11%), age ( 11%) , parkinsonism ( 5 % ), hypothyroidism ( 5%), burns ( 5 %)and benign prostrate hypertrophy ( 5% ). Discussion Blood stream infections are a major challenge in medicine.25 they cause substantial morbidity and mortality. Changing patterns of the isolates, increasing rates of antimicrobial resistance, wide application of new medical technologies like rampant usage of indwelling devices, may change the epidemiology and outcome of BSIs.26 It is therefore important to continually review and update the epidemiology of BSIs mainly with respect to the antibiotic susceptibility pattern of the common pathogens, so that it would be useful for prompt treatment of patients.27 In the present study, an attempt was made to know the etiological profile of clinically suspected BSIs and antibiotic susceptibility pattern of the isolates. The results obtained in the present study were analysed and compared with other studies. JSS Hospital is a tertiary care set up with 235 adults ICU beds and 70% occupancy at any given time. Maximum number of samples were from medicine ICU (30%), medicine ward(18%) followed by Respiratory ICU (16%), Surgery ICU (14%) then Emergency ward (9%), Surgery (4%), ICCU (3%) and Trauma care ward (2%). Similar findings were shown by study of Khaleel et al (2010)28, where majority (49.6%) of positive blood cultures belonged to medicine and allied departments followed by surgical and allied departments (11.3%). Out of the total 100 blood samples received, 64 (64%) were from males and 36 (36%) were from females. Maximum samples obtained were in the age group of 46–60 years, accounting for 37% of the cases. This was followed by age group of 61-75 years (24%). Our study is comparable with the observations made by Mehta et al (2005)10 reported out of 567 blood samples 370 (65%) were males and 197(35%) were females. Out of 100 clinically suspected adult septicaemia cases, blood cultures were positive in 19 (19%) cases whereas in 81 (81%) cases blood cultures were negative. Blood culture positivity in our study was similar to the findings of the study conducted by Arora and Devi et al (2007)24 who reported 20.02% culture positivity. Similar findings (19.3%) by Ayobola et al (2011)7 and (20.5 %) by Garg et al (2007)12 further strengthen our claim. There are many factors which explain variation in blood culture positivity. Firstly, it is difficult to get antibiotic naive patient. Before patient reaches tertiary care centre, some form of antibiotic is already administered.19 Secondly, the variation in blood culture positivity is related to different factors such as the clinical setting, age, selection of patient, specialties, number of blood culture and amount of sample taken.8, 28 Two blood culture samples were collected from two different sites of each patient, to rule out contamination. If two samples from the same patient give positivity with the same isolate, it is considered as the pathogen.In the present study, 100 samples were processed by Bact / Alert 3D system. The automated system issues the blood culture reports earlier than the conventional method. In the present study (table no.1), Gram negative bacilli were found to be the most common cause of adult septicaemia (63%). Gram positive cocci were found in (27%) cases, while fungi (Candida spp.) were isolated in 10% cases.Escherichia coli (27%), Klebsiella pneumoniae (11%), Acinetobacter baumannii (11%), Staphylococcus aureus (10%), CONS (Staphylococcus epidermidis and Staphylococcus hominis) (10%) were the more frequently obtained isolates from adult septicaemia cases. Predominance of Gram negative bacteria were reported by Mehta et al (2005)10 (80.96%), Mehdinejad et al (2009)14 (86.5%). Among Gram positive organisms in our study, Staphylococcus aureus was the commonest (10%) followed by CONS (Staphylococcus epidermidis and Staphylococcus hominis) (10%) and Enterococcus faecalis (6%). This is in accordance to a study carried out earlier by Ayobola et al (2011)7 and Mehta et al(2005)10 who reported Staphylococcus aureus in the percentage of 14.6% and 13.86% of total blood isolates respectively. A high isolation rate of Staphylococcus aureus was reported by Chinna D et al (2013)21 (37.2%) and Anbumani et al (2008)13 (36.4%). Low isolation rate for S. aureus was seen by Barati M et al (2009)29, (6.9%), Garg et al (2007) 12 (8.3%) and Gupta A et al (2012)20 (2.9%). Rising Staphylococcal bacteraemia may have originated from community-acquired infections.30 Such changes in the aetiology at various places are thought to be favoured by geographical location and antibiotic policy advocated in the hospital. This also reflects the better isolation of patients in the hospital and hand washing practices in the ICU or high risk units in the hospital.30, 31 Possibly for similar reasons, cultures in USA and Europe is more likely to reveal gram positive growth.32, 33 Until 1970’s, CONS were mainly recognized as a contaminant. Since then, several studies have reported increasing incidence of infections due to CONS .30, 31 The incidence of CONS was reported as high as 33% of total blood culture tested by Karunakaran et al (2007).6 In some of the larger studies from multidisciplinary hospitals by Falagase ME et al (2006)34 and Lyytikainen O et al (2002)35, coagulase negative staphylococcus (S. epidermidis) was the most common blood culture isolate. With the increased use of aggressive interventions that disrupt the integrity of skin or mucosa and vascular catheters, the likelihood of CONS causing infections is increasing.31 In another study conducted by Karlowsky JA et al9 in U.S. (2002), Out of 82,569 bacterial blood culture isolates, CONS (42%) were the commonest. Others organisms they isolated were S. aureus (16.5%), Enterococcus faecalis (8.3%), E. coli (7.2%), K. pneumoniae (3.6%), Enterococcus faecium (3.5%) and others. However, CONS isolated from culture of patient sample in intensive care settings are more likely to be pathogenic.17 In recent years, CONS have become an important nosocomial pathogen because of the increasing use of medical devices such as long-term indwelling catheters, vascular grafts, and prosthetic heart valves and joints.17 In Our study, Enterococcus faecalis was isolated in 6% of positive blood culture samples which was similar to Mehta et al (2005)10 2.35% and Barati et al (2009)29 1.7%. In contrast Anbumani et al (2008)13 reported a high percentage (4.16) of Enterococci. Similarly, Alam M.S et al (2011)15 reported 6.8% Enterococci in their study. Among gram negative isolates in our study, Escherichia coli (27%) was the commonest followed by Klebsiella pneumoniae (11%) which is similar to a study by Majda Qureshi et al., showing Escherichia coli (16%) as the leading cause followed by Klebsiella pneumoniae(13.3%), Vanitha Rani et al., also showed a percentage of 35.6% for Escherichia coli, but the percentage of Klebsiella pneumoniae (14%) was less. Acinetobacter spp. were isolated in 16 % of positive blood culture samples in our study. Pavani G et al (2013)22 and Latif S et al (2009)16 reported Acinetobacter spp. in 2.5% and 5.1% of samples respectively. In contrast, a very high percentage (32%) of Acinetobacter spp. (mainly A. lwoffii followed by A. baumannii) were reported by Barati M et al (2009)29 at a university hospital in Iran. The high percentage of this organism in their study can be explained by the fact that they included children and critically ill patients. Acinetobacter spp. were reported in 13.6% of blood isolates by Elouennass M et al (2008).36 Acinetobacter spp. has been increasingly implicated as a cause of a wide spectrum of infections including community and hospital acquired infections associated with intravenous catheters and contaminated respiratory therapy equipment among patients with impaired host defences in intensive care units37, 38.In the culture study, Pseudomonas aeruginosa was isolated in 5% of positive blood culture samples. Similar observations were made by Qureshi M et al (2011)18, Arora et al (2007)24 and Asghar A.H.et al (2006)11 who reported this organism in 10.7%, 7.63% and 9.8% of positive blood culture samples. A high percentage of isolation of Pseudomonas aeruginosa were reported by Mehta et al10 (19.75%) and by Garg et al (2007)12 (16%). Abbreviations: CONS - coagulase-negative staphylococci; S.aureus- Staphylococcus aureus; E. coli -Escherichia coli; K. pneumoniae - Klebsiella pneumoniae, Streptococcus viridans group E. faecium - Enterococcus faecium, C. albicans-Candida albicans. Fluit et al (2000)39 and Diekema et al (2000)40 reported that S. aureus and E.coli were the most frequently isolated organisms from hospitalized patients in the United States and Europe. Apart from Gram positive and Gram negative organisms, Candida spp. were isolated in two positive blood culture samples. The isolates were Candida glabrata(5%) and Candida tropicalis(5%). In a study conducted Gupta A et al (2012)20 reported high percentage of fungal isolates (13%), mainly Candida spp. of which non albicans Candida such as Candida tropicalis, Candida glabrata, Candida krusei predominated, this observation is also supported by Akbar DH et al(2001) and Diekema DJ et al(2002), Pfaller MA et al (1998). Latif S et al (2009) 16 reported that of their blood isolates, 2.4 % were Candida albicans. Candida blood stream infections are on rise because of high usage of antibiotics and various other co-morbidities. Although albicans remains predominant species involved, non albicans Candida are also being recognized as important pathogen and likely to replace albicans as predominant pathogen.6 Usually BSIs are monomicrobial in nature. Polymicrobial growth in blood culture indicates either contamination or severe infection or infection at multiple sites.19, 28 The rate of Polymicrobial BSI as indicated by isolation of >2 organism varies between 1 to 18%.5,12,28 Usually polymicrobial infections are hospital acquired and seen in immunocompromised patient.12 In our study all blood culture yielded single organism. Present study, shows different medical specialties and bacteriological profile in these. Maximum number of samples were from medicine ICU (30%), medicine ward(18%) followed by Respiratory ICU (16%), Surgery ICU (14%) then Emergency ward (9%), Surgery (4%), ICCU (3%) and Trauma care ward (2%). Escherichia coli was (27%) predominant gram-negative organism in ICU which was followed by Klebsiella pneumoniae (11%). Among gram positive organisms, Staphylococcus aureus (10%) was predominantly present followed by Coagulase negative Staphylococcus (10%). In present study (table no.2), genito-urinary infections (79%) followed by respiratory infections (53%), metabolic disorders (53%), skin and soft tissue infections (11%) were the common clinically suspected primary sources of infection. Similar to the results of our study, Siegmanigra Y et al (2002)41 also showed that urinary tract infection (associated with 39% of all bacteraemia) was the most common source of bacteraemia. This was followed by primary bacteraemia (vascular device associated and endovascular infections etc) which contributed to 17% of the episodes of bacteraemia. McDonald JR et al (2005)42 in their study documented that 23.8% of the study patients had urinary tract infections and 17.2% had pneumonia. Primary bacteraemia was seen in 29.6% patients. Son JS et al (2010)47 found that in CA-BSI, Urinary tract infections 31.3% (119/380) was the main source of infection and that primary bacteraemia was the main source of infection in HA-BSI. Antimicrobial resistance The increase in the number of isolates showing resistance to at least 3 drugs is alarming. This phenomenon is termed as multidrug resistance. In our study most of the isolates obtained were multi-drug resistant. The problem of antibiotic resistance was recognized and reported in the 1980’s where multiple resistant strains were seen.43 Moniri et al (2006)44 showed that 51 out of 69 (73.9%) of their isolates were resistant to three or more antimicrobials. Japoni et al (2008)45 recorded the occurrence of multi-drug resistance. Their blood isolates were resistant to 4 antimicrobial agents. Edoh and Alomatu et al (2007)43 also reported a resistance to 3 antibiotics by all isolates. The incidence of fungal blood stream infections has reportedly increased with rates of 5.4% in 1980 and 9.9% in 1990, as reported by the National Nosocomial Infection surveillance System for the United States Hospitals.46 In present study (table no.6), Isolated fungi- Candida glabrata and Candida tropicalis were sensitive to fluconazole, voriconazoleand flucytosine. Candida tropicalis isolate was resistant to amphotericin B. Trends towards increasing numbers of non-albicans Candida among bloodstream pathogens have also been reported.46 The preponderance of non-albicans Candidemia is important to note, as empirical therapy with fluconazole, while appropriate against C. albicans, may not cover other species of Candida. In the present study Diabetes Mellitus was identified as the major risk factor (37%). The other risk factors were intravascular catheterization (32%), VAP (16%), Carcinoma/ CLL, Age, Dialysis (11%), Parkinsonism, Long Steroid Therapy, Hypothyroidism, Burns, BPH (5%). Coinciding with our study, Michalia M et al (2009) reported Diabetes mellitus as an independent risk factor for ICU – acquired bloodstream infections; Fram D et al (2015) reported dialysis as a risk factor blood stream infection. And Urlich Seybold et al(2004)., concluded that extremes of age, long term care facility, urinary catheters, hospitalisation and any form of dialysis as a risk factor for BSI. Detection of bloodstream infections is one of the most important tasks performed by the clinical microbiology laboratory. Rapid identification results and antimicrobial susceptibility tests are essential for guiding clinicians in the selection of the most appropriate treatment for patients with bloodstream infections. The Vitek 2 system (bio-Me´rieux, France) was introduced to the market in 1999, and its ability to identify and determine the susceptibility of both gram-positive cocci and gram-negative rods has been evaluated in several reports. Ability and accuracy of Vitek 2 system has been well studied in comparison with other automated systems and reference methods. This is supported by Barenfanger, J., et al (1999), Barry, J et al (2003). Evangelista, A(2002). Funke, G.(1998).
Summary
CONCLUSION
REFERENCES
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Home
Home