|
Table of Content Volume 15 Issue 3 - September 2020
Prevalence and antibiotic susceptibility pattern of proteus species from various clinical samples coming to a tertiary care hospital, Rajkot
Nirali Daftary1*, Dipika Patel2
1Senior Resident, P.D.U Medical College, Rajkot, INDIA. 2Tutor, Government Medical College, Surat, Western, 75/B, Dharanagar Society, Abrama, Valsad-396002. INDIA. Email: nirali.daftary10@gmail.com
Abstract Background: Proteus species are a well known group of bacteria which cause various infections. However in many laboratories in the developing countries, differentiation of the genus Proteus into species is not generally done during bacteriological diagnosis due to high cost and special skills involved. Aims and Objectives: To study the prevalence of different Proteus species at a tertiary care hospital, Rajkot. To determine the antibiotic susceptibility pattern of various Proteus species. Settings and design: This is an observational cross-sectional study Materials and Method: All the 5895 samples were collected from various sites (pus, urine, blood etc.)and were subjected to routine microscopy. After that they were cultured on blood agar, MacConkey agar and Nutrient Agar and the colony characteristics like swarming etc were observed. Various biochemical tests were done to identify the species of various Proteus bacteria. Then the antibiotic susceptibility test was done using Kirby Bauer’s method of disc diffusion. Results: 186 of the 5895 clinical samples showed growth of proteus species of which P.mirabilis was highest at 88.7% followed by P.vulgaris at 9.13%. Male surgical ward had the highest prevalence at 41.93% and Swine Flu and Plastic ward at 0.53%. Male predominance was observed and the age group of 51-60 years showed maximum growth of Proteus spp at 23.6%. All samples were susceptible to Meropenem at 100% and least susceptible to ampicillin at 0.53%. Extended Spectrum Beta Lactamase production was seen more in P.vulgaris at 35.29% than in P.mirabilis at 21.81%. Conclusion: Proteus was isolated from 3.15% of all the clinical samples. Most isolates were from pus samples. The antibiotic resistance was also high amongst them. Proper aseptic care and judicial antibiotic usage will help in reduction of proteus infection. Key Word: proteus species.
INTRODUCTION Proteus species are among the commonly implicated pathogens in hospital as well as community acquired infections. This pathogen has a diverse mode of transmission and hence can cause infection in different anatomical sites of the body. Some of the incriminating sources of transmission are soil, contaminated water, food, equipments, intravenous solutions etc. There are reports of 2% to 13.4% prevalence rates of proteus infections in the tertiary care hospital. Different species of proteus are encountered in human infections; however, bacteriological diagnosis up to identification of species is rare in many laboratories due to the cost and special skills involved. There is therefore limited documented information relating to patients’ demographics and antibiotic susceptibility levels for infections caused by the various species of proteus. This study seeks to determine the prevalence of the various proteus infections in relation to patients’ demographics and the response of the different species to commonly prescribed antibiotics at a tertiary care hospital, Rajkot.
MATERIALS AND METHODS Sample Collection: Different clinical samples such as pus, ear swabs, purulent material from wounds or abscesses, urine, sputum, blood or aspirated of cerebrospinal fluid, ascetic fluid etc collected from 5895 patients were cultured to isolate the organisms. Demographic data (such as age, sex, in-patient and out-patient status) of the patients was recorded prior to sample collection. Cultivation and Identification: The clinical samples were collected and their routine microscopy including Gram stain and motility was done. Then they were aseptically inoculated on plates of MacConkey agar, Nutrient agar, blood agar and incubated at 37oC for 24 hours. The morphological characteristics of the colonies including size, shape, colour, swarming, pigmentation and haemolytic nature were recorded. Pale colonies were first tested by oxidase test and catalase test. Then their motility and gram stain was done. Suspected proteus colonies were isolated and identified through biochemical tests based on phenyl pyruvate production, H2S gas production (positive in proteus and negative in Providencia and morganella), methyl red and urease reactions (positive in proteus and morganella; negative in Providencia spp), citrate utilization (positive in proteus and providencia and negative in morganella), glucose fermentation. Indole production differentiated P. vulgaris isolates from the other species. Ornitihine is positive only in Morganella. Maltose fermentation is positive in P. vulgaris and negative in P.mirabilis.
Antimicrobial susceptibility tests Modified Kirby-Bauer disk diffusion method (Cheesebrough, 2000) was used to test the susceptibility of the proteus isolates to different antimicrobial agents: Meropenem, Pipercillin/tazobactam, Ceftazidime/sulbactam, cefepime, ceftazidime, cefotaxime, cefuroxime, Ampicillin , ampicillin/sulbactam, amikacin, gentamicin , cotrimoxazole . The inocula were prepared by growing the various proteus species on separate agar plates and colonies from the plate were transferred with inoculating loop into 3 ml of normal saline in a test tube. The density of these suspensions was adjusted to 0.5 MacFarland standards. The surface of Muller-Hinton agar plate was evenly inoculated with the organisms using a sterile swab. The swab was dipped into the suspension and pressed against the side of the test tube to remove excess fluid. The wet swab was then used to inoculate the Muller-Hinton agar by evenly streaking across the surface. Then the antibiotic discs were applied to the surface of the inoculated agar and the plates were incubated overnight at at 37oC for 24 hours. The diameter of the zone of growth-inhibition observed was measured and compared to the chart provided by Clinical Laboratory Standards Institute.
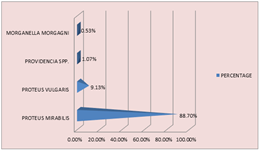
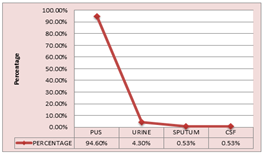
RESULT AND DISCUSSION 186 Proteus species were recovered from 5895 clinical samples and the prevalence rate was 3.15. Proteus mirabilis was the highest with 88.7%, followed by Proteus vulgaris 9.13%, Providencia spp being 1.07% and least was morganella morgagni at 0.53%. Pus samples contributed to the highest percentage of Proteus at 94.6%, followed by urine at 4.3%, then sputum 0.53% and cerebrospinal fluid 0.53%.
Table 1: Species wise distribution
Figure 1: SPECIES DISTRIBUTION
Table-2: Sample wise Distribution
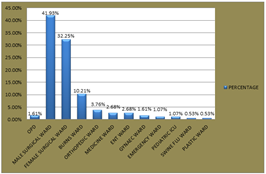
Figure 2: TYPE OF SAMPLE There was a male predominance of 55% over females 45%.The male surgical ward had the highest prevalence at 41.9%, followed by female surgical ward at 32.5% and the least in swine flu ward and plastic ward at 0.53% each whereas the outdoor patient department had 1.61%. All age groups except 81-90 years age group had atleast one species present with maximum prevalence in the 51-60 years age group at 23.6% and the least in 1-10years at 2.15%. Table 3: Type of admission
Figure 3: TYPE OF ADMISSION Other types of samples and their bacterial growth were as follows:
Table 4: Total samples and their categorization
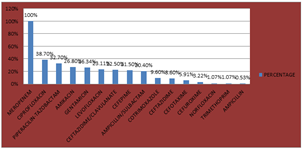
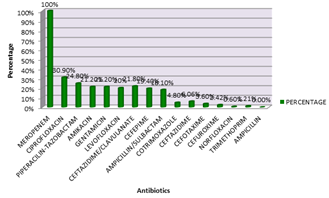
Anitmicrobial susceptibility of proteus isolates The Proteus isolates recovered were highly susceptible to meropenem at 100%. However 38.7% were susceptible to ciprofloxacin, 32.7% to piperacillin-tazobactam, 26.8% to amikacin, 26.34% to gentamicin, 42 samples showed extended spectrum beta lactamase resistance (ESBL), 21.5% were susceptible to cefepime, 8.6% to ceftazidime, 5.91% to cefotaxime, 3.22% to cefuroxime, 1.07% to norfloxacin, 1.07% to trimethoprim. ESBL was seen more in P.vulgaris at 35.29% than in P.mirabilis at 21.81%.
Table 5: Antibiotic resistance pattern in Proteus spp
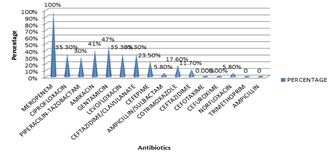
Figure 4: ANTIBIOTIC RESISTANCE of Proteus species P.mirabilis was also most susceptible to Meropenem at 100% and least susceptible to norfloxacin at 0.6%. Table 6: Antibiotic resistance pattern in Proteus mirabilis
Figure 5: ANTIBIOTIC RESISTANCE IN P. MIRABILIS
P. vulgaris was also most susceptible to Meropenem at 100% and least susceptible to ampcillin/sulbactam and norfloxacin at 0.6%
Table 7: Antibiotic Resistance Pattern in Proteus vulgaris
Figure 6: ANTIBIOTIC RESISTANCE IN P. VULGARIS
DISCUSSION Species identification and surveillance of antimicrobial resistance is essential in management and control of infections. These practices are usually absent in many hospitals mainly due to costs involved. In this study we investigated the presence of Proteus species from 5895 clinical samples collected from January 2019 to December 2019. Four proteus species were identified to be responsible for causing infections in various anatomical sites. P. mirabilis was the most common species isolated, accounting for 88.7% of all infections hence responsible for the majority of Proteus infections. This result agrees with similar studies conducted in England, wales and Northern Ireland [Chow, 1979; Jones, 2003]. Pus recorded the highest percentage of proteus isolates 94.6%, followed by urine 4.3%. Proteus therefore is the common cause of wound infections in Rajkot and other parts of Gujarat, India. Our finding thus partially supports the findings of those from Europe and Asia [Reslinki, 2005; Chung, 1999] which showed Proteus species to be more commonly encountered in pus than in other clinical samples. According to our study maximum infection in urine sample was of P.mirabilis which is in congruation with the finding which supports that P.vulgaris and P.penneri infections of the urinary tract are rare (Chung, 1999; Foxman, 2000; Naxal, 1994) whereas P.mirabilis has a higher propensity for colonizing the urinary tract due to difference in its pathogenicity (mobley,1994). Proteus infections were also common among the in-patients (98.3%) as compared to out-patients (1.61%). Out of the 186 clinical specimens from whom Proteus was recovered, 55% were collected from males and 45% from females. The study showed a significant difference between the males and females infected with Proteus. The Proteus infections were detected in all age groups from 1 to 99 years where 51-60 years age group registering as the highest group infected (23.6%). The Proteus species isolated were found to have high antimicrobial resistance against third generation of Cephalosporin antibiotics. All the proteus species showed multidrug resistance. 22.5% of the total clinical samples were ESBL producers. ESBL was seen more in P.vulgaris at 35.29% than in P.mirabilis at 21.81%.The high antibiotic resistance of proteus may be an indication of the resistance levels among the enterobacteriaceae and perhaps salmonella since the indiscriminate ingestion of antibiotics provides selective pressure, leading to a higher prevalence of resistant bacteria (Levy, 1999) which is very common in developing countries like India. Not only are these species potential causes of infections but also potential reservoirs of resistance genes that could be transferred to other bacterial pathogens. The high levels of B-lactamase production and multi-drug resistance of the isolates are indications of an increase in the resistance menace reported by earlier studies (Newman, 2006). P.mirabilis, P. vulgaris, Providencia an Morganella morgagnii are the species implicated in Proteus infections; wounds recorded the highest incidence of Proteus infections at a tertiary care hospital, Rajkot. The species were susceptible to Meropenem, Ciprofloxacin, Levofloxacin. They were however resistant to ampicillin, cefotaxime, cefuroxime hence these don’t form part of the empirical antibiotics for the treatment of Proteus infections at the tertiary care hospital. B-lactamase production and multi-drug resistance have all been exhibited by the isolates. This study is therefore a step towards the generation of national data on the prevalence of antimicrobial resistant pathogens. Acknowledgement We are grateful to the co-operation and the technical help at the tertiary care hospital, Rajkot.
REFERENCES
Policy for Articles with Open Access
|
|
 Home
Home