|
Table of Content - Volume 19 Issue 2 - August 2021
Detection of mrkD gene in clinical isolates of biofilm producing Klebsiella pneumoniae
Preethi Chilupuri1, Sawjanya Gudisi2*, K Prakash3
1Assistant Professor, 2Assistant Professor, 3Professor, Department of Microbiology, Chalmeda Anand Rao Institute of Medical Sciences, Karimnagar, INDIA. Email: sgurnule28@gmail.com
Abstract Background: Isolates of Klebsiella pneumoniae are responsible for opportunistic infections in humans, particularly of the urinary, respiratory tracts. These bacteria express type 3 fimbriae that have been implicated in binding to eukaryotic cells and matrix proteins. They are often found in a variety of environmental niches in aggregates called biofilms. The development of biofilms by pathogenic bacteria is believed to play an important role in facilitating evasion of host defence mechanisms, communication between bacterial cells and protection against antibiotic action. Materials and methods: 52 Klebsiella pneumoniae isolated from different clinical samples were studied for biofilm production by tube method and congo red agar method. PCR for detection of mrkD gene was done in biofilm producers. Results: In 52 isolates 38 (73.07%) are positive for biofilm production by tube method and 35 (67.30%) are biofilm producers by CRA method. Out of the 28 biofilm producers mrkD gene is detected only in 8(21.08%). Conclusions: various studies have revealed that type 3 fimbriae are important in K. pneumonia biofilm formation. However, there are several genes coding for fimbriae each of them determining specific functions in virulence and biofilm formation. Keywords: Klebsiella pneumoniae, biofilm, mrkD gene.
INTRODUCTION Klebsiella pneumonia was isolated for the first time in 1882 by Friedlander from the lungs of patients who died after pneumonia. This encapsulated bacterium, initially named Friedlander’s bacillus was renamed Klebsiella in 1886. It is a Gram-negative bacterium belonging to the Enterobacteriacea family, often found in a variety of environmental niches. Due to its high pathogenicity, K. pneumonia, in the pre-antibiotic era, was considered as an important causative agent of community-acquired (CA) infections and severe forms of pneumonia, especially in alcoholics and in diabetic patients. The greater adhesiveness and presumably also the invasiveness of strains may play an important role in the recurrent infections and able to persist despite appropriate antibiotic treatment.1 Starting from the early 1970s, K. pneumoniae epidemiology and its spectrum of infections significantly changed when this microorganism was established in the hospital environment and became a leading cause of nosocomial infections. Considerable efficiency of colonization, accompanied by acquired resistance to antibiotics, has enabled it to persist and spread rapidly in healthcare settings causing the urinary tract, wounds, lungs, abdominal cavity, intra-vascular devices, surgical sites, soft tissues infection and subsequent bacteremia. K. pneumoniae was reported to be able to grow in vitro as a biofilm since the end of the 1980s, but clear evidence of an in vivo biofilm was provided only in 1992 by Reid and co-workers. Biofilms are currently defined as structured bacterial communities enclosed in a self-produced exopolysaccharide matrix and adherent to abiotic or biological surfaces. Its formation can be divided into distinct stages, from the initial attachment of bacteria to the surface to the formation of mature biofilm with a characteristic three-dimensional architecture. Biofilm producing bacteria are responsible for many recalcitrant infections and are notoriously difficult to eradicate. They exhibit resistance to antibiotics by various methods like restricted penetration, decreased growth rate and expression of resistance genes.2 Type 1 or type 3 fimbrae, as well as the capsule and the LPS are the virulence factors mostly involved in the ability of biofilm formation. The expression of type 3 fimbriae has been shown to promote biofilm formation. In particular, growth of K. pneumoniae on abiotic surfaces is facilitated by the MrkA type 3 fimbrial protein, whereas growth on surfaces coated with a human extracellular matrix (HECM) requires the presence of the type 3 fimbrial adhesion MrkD.
MATERIALS AND METHODS Prospective study conducted in Department of Microbiology, Mamata Medical College, Khammam during May 2013 to March 2014. A total of 52 clinical isolates of Klebsiella pneumonia isolated from urine, sputum, pus from infected wounds, infected devices and skin surfaces were investigated in this study. Isolates were initially identified by standard microbiological techniques including Gram stain, hanging drop, sugar fermentation and biochemical tests. Detection of biofilm formation:
A qualitative assessment of biofilm formation was determined as previously described by Christensen et al.14 TSBglu (10mL) was inoculated with loopful of microorganism from overnight culture plates and incubated for 24 hours at 37°C. The tubes were decanted and washed with PBS (pH 7.3) and dried.dried tubes were stained with crystal violet (0.1%). Excess stain was removed and tubes were washed with deionized water. Tubes were than dried in inverted position and observed for biofilm formation. Biofilm formation was considered positive when a visible film lined the wall and bottom of the tube. Ring formation at the liquid interface was not indicative of biofilm formation. Experiments were performed in triplicate and repeated three times.
Freeman et al. have described a simple qualitative method to detect biofilm production by using Congo Red Agar (CRA) medium. CRA medium was prepared with brain heart infusion broth 37 g/L, sucrose 50 g/L, agar No. 1 10 g/L and Congo Red indicator 0.8 g/L. First Congo Red stain was prepared as a concentrated aqueous solution and autoclaved (121 oC for 15 minutes) separately from the other medium constituents. Then it was added to the autoclaved brain heart infusion agar with sucrose at 55 oC. CRA plates were inoculated with test organisms and incubated at 37 oC for 24 h aerobically. Positive result was indicated by black colonies with a dry crystalline consistency. The experiment was performed in triplicate and repeated three times.
Bacteria were grown overnight with shaking at 37°C in 10 ml of LB. Briefly, 1 ml of the bacterial suspension was centrifuged (13,000 × g, 5 min), the pellet was recovered and resuspended in 300 μL of sterile water and heated to 90°C for 10 min. After centrifugation (13,000 × g, 5 min), the supernatant was directly used as template DNA for PCR amplification. PCR amplifications were performed using the Expand Long Template PCR system (Roche, Mannheim, Germany). In a final volume of 50 μM, 5 μL of template DNA was added to the amplification solution containing 5 μl of 10 x kit buffer, 0.5 μl of Taq polymerase, each deoxynucleoside triphosphate at a final volume of 0.1 μl, and each primer at a final concentration of 25 μM. Two primers were used: MrkD F: 5’- CCACCAACTATTCCCTCGAA-3’ and MrkD R: 5’-ATGGAACCCACATCGACATT-3’. Cycling conditions were as follows: one denaturation step of 2 min at 94°C and 10 initial cycles of 10 s at 94°C, 30 s at 63°C, and 15 min at 68°C, followed by 20 iterative cycles of 10 s at 94°C, 30 s at 63°C, and 15 min plus 20 s for each new cycle at 72°C. A final elongation step of 7 min at 72°C was added. Amplified products were examined by 1.3 % agarose gel electrophoresis in 0.5 x TBE buffer.
RESULTS In 52 clinical isolates of Klebsiella pneumoniae 38 (73.07%) are positive for biofilm production by tube method and 35 (67.30%) are biofilm producers by CRA method.
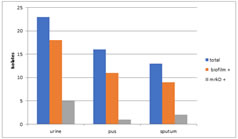
Graph 1: Distribution of biofilm production by both the methods In 52 clinical isolates from which Klebsiella pneumonia is isolated 18 in 23 urine, 11 in 16 pus and 9 in 13 sputum samples are biofilm producers. 5 in 18 urine, 1in 11 pus and 2 in 9 sputum samples with biofilm producing Klebsiella pneumonia are positive for mrkD gene on PCR. Table 1: Table showing the results
Graph 2: Graphical representation of results
Figure 1: Black Colonies on blood agar with congo red supplement
Figure 2: Tube Test for presence of fimbriae in klebsiella species.
Figure 3: Polymerise chain Reaction Test
DISCUSSION Although K. pneumonia is a leading cause of Gram-negative nosocomial infections and is associated with a high mortality rate not much is known about its pathogenic scheme beyond the role of capsule. In the established paradigm of K. pneumonia pathogenesis, the anti-phagocytic polysaccharide capsule is the most distinguishing characteristic and most studied virulence factor. Only a few other factors including LPS and pili have been proposed as virulence factors, but their contribution to host pathogenesis has not been examined in detail. The ability of these bacteria to associate into communities in biofilms is central to their pathogenicity as they confer protection from bactericidal molecules present on host tissues.3 Over the past few decades, K. pneumonia has emerged as a significant cause of severe nosocomial infections due to it being difficult to treat. The persistence of this species in hospitals is partly due to its ability to adhere and multiply on inanimate surfaces. Little is known about the bacterial factors involved in K. pneumoniae adherence to abiotic surfaces (Podschun and Ullmann, 1998).1 It has been previously reported that quorum sensing is involved in biofilm formation by K. pneumonia (Balestrino et al., 2005).2 Other studies demonstrated the influence of capsule and extended-spectrum beta-lactamase encoding plasmids upon K. pneumonia adhesion (Hennequin and Forestier, 2007).3 A high prevalence of type 1 and type 3 pili has been previously observed in clinical and environmental K. pneumonia isolates (Livrelli et al., 1996; Martynenko et al., 1992; Podschun et al., 2001; Schurtz et al., 1994).4-7 Type 3 pili, mostly found in K. pneumonia are also involved in biofilm formation by this bacterial species (Di Martino et al.,)8 and are generally considered as a virulence factor. In the present study 52 strains of K. pneumonia were isolated from different clinical samples i.e., urine, pus and sputum. Biofilm formation was identified by two methods. 38 (73.07%) in total 52 were positive for biofilm production by tube method and 35 (67.30%) were biofilm producers by CRA method.
CONCLUSION Present study revealed that type 3 fimbriae are important in K. pneumonia biofilm formation. However, there are several genes coding for fimbriae each of them determining specific functions in virulence and biofilm formation
RERERENCES
Policy for Articles with Open Access
|
|
 Home
Home